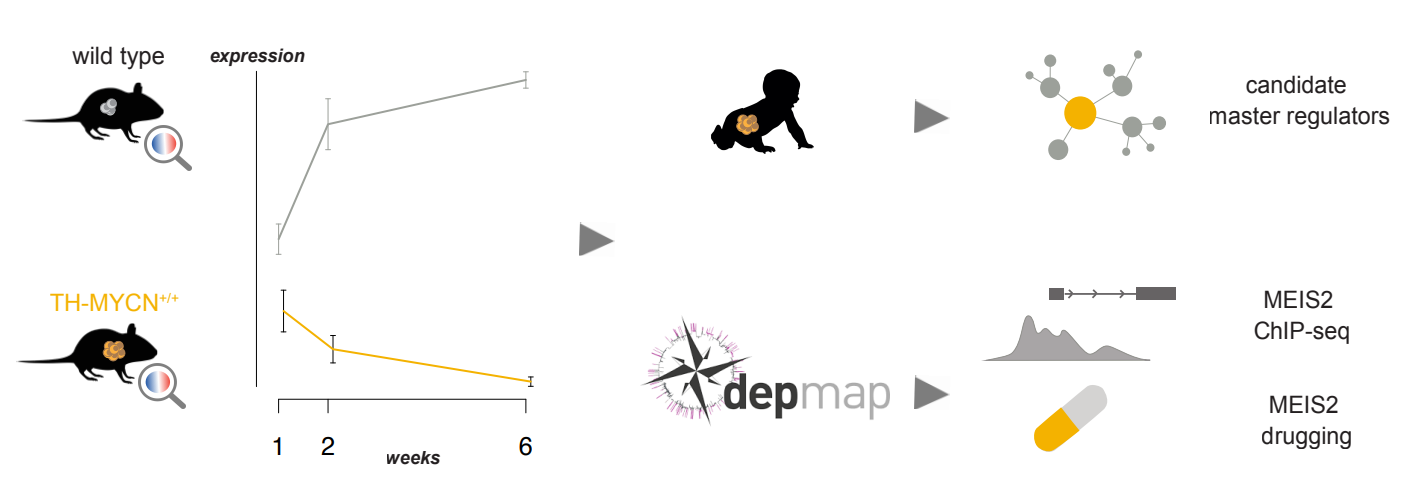
Our discovery efforts focused on identifying cooperating interactors and vulnerabilities in the MYCN regulatory network. MYCN-driven neuroblastomas can be modeled in mice with morphologic and genomic features that recapitulate human disease. We performed a time-resolved analysis of the dynamic transcriptional changes of protein coding genes during murine TH-MYCN driven neuroblastoma development, focusing on timepoints representing tumor initiation and early tumor growth. Amongst the top up-regulated genes, we identified gm6890, a component of the highly evolutionary conserved KEOPS protein complex and associated with high expression levels in primary neuroblastoma being associated with poor survival. Next, we triangulated expression changes of key genes with publicly available exome-wide CRISPR-cas9 knockout analyses on a panel of human neuroblastoma cell lines and patient survival data. This unique data resource uncovered the relevance of MEIS2 as putative early cooperating initiating factor for neuroblastoma. Analysis of the genome-wide binding profile of MEIS2 in MYCN-amplified neuroblastoma cell lines showed a striking overlap with enhancer-driven gene expression in regions of open chromatin, providing evidence that MEIS2 is a novel member of the adrenergic neuroblastoma core-regulatory circuitry. Finally, cross-species integrative transcriptomics approach with expression data of 498 primary human neuroblastomas allowed to identify transcription factors acting as master regulators with FOXM1 as top ranked gene.